Wieden Group
Email: hj.wieden@uleth.ca
Research
Synthetic biology focuses on the rational design of biological systems using a multidisciplinary approach bridging classical molecular biology method and sophisticated bioinformatics and systems biology approaches. Our work focuses on the development of novel engineered expression platforms enabling innovative application addressing environmental and medical issues as well biotechnology.
Within the Alberta RNA Research and Training Institute (ARRTI), we belong to the the Laboratory for biomolecular design and engineering and contribute to the Laboratory for Synthetic Biology.
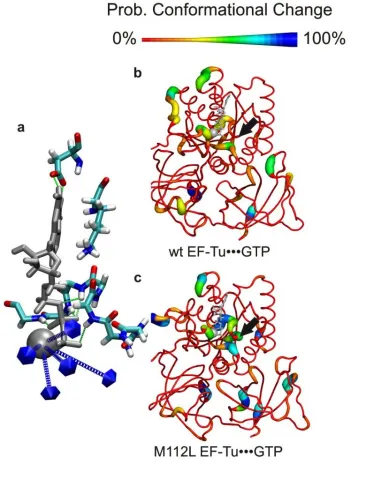
Biomolecules such as Proteins and RNAs are intrinsically flexible, and typically undergo a wide variety of motions at normal temperatures. X-rays crystallographic structures as well as Cryoelectron microscopic studies of these biomolecules demonstrated that a high degree of conformational flexibility is important for the function. The flexibility and dynamics of proteins such as elongation factors has been optimized by evolution for their activities and functions. In order to analyze the role of the dynamical properties on their function, and to bridge the gap between the static structural data and the huge amount of biochemical and kinetic information that is available, the conformational flexibility of elongation factors such as EF-Tu and EF-G is studied using computational methods such molecular dynamics simulation in conjunction with advanced biophysical approaches including single molecule methods.
Molecular Mechanism of Antibiotics
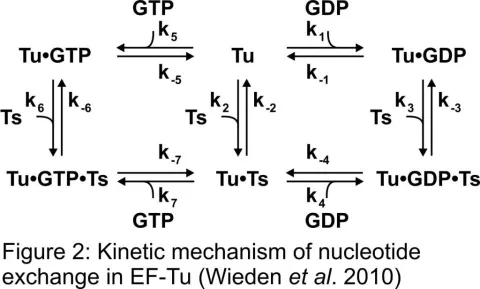
With the steady emergence and spread of antibiotic resistant pathogens, the development of new antibiotics is increasingly important. We study the molecular mechanisms of antibiotic function using a multidisciplinary approach based on advance biophysical methods in order to provide a framework for the development of novel antimicrobial strategies. Our research focuses on antibiotics that target the cellular machinery of the pathogen that is responsible for translating genetic information into functional proteins, a process called translation. The detailed understanding of the involved processes is of fundamental importance for the development of new types of antibiotics. We study the molecular requirements for the inhibition of translation and analyze how resistance mechanisms work. On going research contributes to the development of novel tests that will allow us to search for chemical compounds that will effectively inhibit translation.
Selected Publications
- Coatham, M.L., Brandon, H.E., Fischer, J.J., Schummer, T., and Wieden, H.-J. (2016) The conserved GTPase HflX is a ribosome splitting factor that binds to the E-site of the bacterial ribosome. Nucleic Acids Res.DOI: 10.1093/nar/gkv1524 (IF: 9.11)
- Brandon, H.E., Friedt, J.R., Glaister, G.D., Kharey, S.K., Smith, D.D., Stinson Z.K., and Wieden, H.-J. (2015) A New Part Class Utilizing Programmed Ribosomal Frameshifts. Translation DOI: 10.1080/21690731.2015.1112458
- Rosler, K.S., Mercier, E., Andrews, I.C., and Wieden, H.-J., (2015) Histidine 114 is critical for ATP hydrolysis by the universally conserved ATPase YchF. J. Biol. Chem. 290, 18650-18661. DOI: 10.1074/jbc.M114.598227 (IF: 4.60)
- Tillault, A.S., Fourmann, J.B., Loegler, C., Wieden, H.-J., Kothe, U., and Charpentier, B., (2015) Contribution of two conserved histidines to the dual activity of archaeal RNA guide-dependent and -independent pseudouridine synthase Cbf5. RNA 21, 1233-1239 DOI: 10.1261/rna.051425.115 (IF: 4.622)
- De Laurentiis, E.I., and Wieden, H.-J., (2015) Identification of two structural elements important for ribosome stimulated GTPase activity of Elongation Factor 4 (EF4/LepA). Scientific Reports5, 8573 DOI: 10.1038/srep08573 (IF: 5.578)
- Mercier, E., Girodat, D., Wieden, H.-J. (2015) A conserved P-loop anchor limits the structural dynamics that mediate nucleotide dissociation of EF-Tu. Scientific Reports 5, 7677 DOI: 10.1038/srep07677 (IF: 5.578)
- Boël, G., Smith, P.C., Ning, W., Englander, M.T., Chen, B., Hashem, Y., Testa, A.J., Fischer, J.J., Wieden, H.-J., Frank, J., Gonzalez, R.L. Jr., Hunt, J.F. (2014) The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat. Struct. Mol. Biol. 21, 143-151. (IF: 11.633)
- Friedt, J., Leavens, F.M., Mercier, E., Wieden, H.-J., Kothe, U. (2014) An arginine-aspartate network in the active site of bacterial TruB is critical for catalyzing pseudouridine formation. Nucleic Acids Res. 42, 3857-3870. (IF: 8.808)
- Becker, M., Gzyl, K., Altamirano A.M., Vuong, A., Urban, K., Wieden, H.-J. (2012) The 70S ribosome modulates the ATPase activity of Escherichia coli YchF. RNA Biology 9(10): 1288-1301. (IF 5.377)
- Fischer, J., Coatham, M.L., Eagle Bear, S., Brandon, H.E., De Laurentiis, E.I., Shields, M.J., Wieden, H.-J. (2012) The ribosome modulates the structural dynamics of the conserved GTPase HflX and triggers tight nucleotide binding. Biochimie 94(8):1647-1659. (IF: 3.123)
- Gruninger R.J., Dobing, S., Smith, A.D., Bruder LM, Selinger, L.B., Wieden, H.-J., Mosimann, S.C. (2012) Substrate binding in protein tyrosine phosphatase-like inositol polyphosphatases. J. Biol. Chem. 287, 9722-9730. (IF: 4.6)
- Delaurentiis, E.I., Mo, F., and Wieden, H.-J. (2011) Construction of a fully active Cys-less Elongation Factor Tu: Functional role of the conserved Cystein 81. Biochim. Biophys. Acta. 1814, 684-692. (IF: 3.191)
- Wieden, H.-J., Mercier, E., Gray, J., Steed, B., Yawney, D. (2010) A combined molecular dynamics and rapid kinetics approach to identify conserved 3 dimensional communication networks in Elongation Factor Tu. Biophys J. 99, 3735-3743 (IF: 3.832)
- Shields, M., Fischer, J., Wieden, H.-J. (2009) Toward understanding the function of the universally conserved GTPase HflX from Escherichia coli: A kinetic approach. Biochemistry 48, 10793-10802 (IF: 3.194)
Group Members (Fall 2016)
- Dr. Rajashekhar Kamalampeta (PDF)
- Dr. Senthil Kumar Duraikannu Kailasam (PDF)
- Dr. Andrew Hudson (PDF)
- Harland Brandon (PhD Student)
- Dylan Girodat (PhD Student)
- Luc Roberts (PhD Student)
- Taylor Sheahan (PhD Student)
- Katherine Gzyl (MSc Student)
- Dustin Smith (MSc Student)
- Justin Vigar (MSc Student)
- Rhys Hakstol (MSc Student)
- Dora Capatos (MSc Student)
- Jalyce Heller (MSc Student)
- Cristina Carvalho Barbosa de Souza (MSc Student)
- Davinder Kaur (MSc Student)
- Darren Gemmill (Undergraduate)
- Chelsi Harvey (Undergraduate)
- Dr. Binod Pageni (Research Associate)
- Fan Mo (Research Associate)
- Emily Wilton (Scientific Assistant)
Research Funding (Current)




